IMU-frame to body-frame transformations#
Consider loading the logging module and setting up a logger to
monitor progress:
1# Set up
2import importlib.resources as rsrc
3import numpy as np
4import pandas as pd
5import matplotlib.pyplot as plt
6import skdiveMove.imutools as imutools
7from scipy.spatial.transform import Rotation as R
8from skdiveMove.imutools.imu import (_ACCEL_NAME,
9 _OMEGA_NAME,
10 _MAGNT_NAME)
11from skdiveMove.imutools.imu2body import (_LEG3X1_ANCHOR,
12 _euler_ctr2body)
13
14_FIG1X1 = (11, 5)
15_FIG3D1X1 = (11, 8)
16_FIG3X1 = (11, 11)
17_FIG4X1 = (10, 10)
18
19pd.set_option("display.precision", 3)
20np.set_printoptions(precision=3, sign="+")
21%matplotlib inline
Load saved pre-processed (properly calibrated and in a right-handed, front-left-up frame) data:
22icdf = (rsrc.files("skdiveMove") / "tests" / "data" /
23 "gertrude" / "gert_imu_frame.nc")
24icsv = (rsrc.files("skdiveMove") / "tests" / "data" /
25 "gertrude" / "gert_long_srfc.csv")
Set up an instance to use throughout:
26# If data are in NetCDF and CSV files, one can take advantage of the
27# class method `IMU2Body.from_csv_nc` to instantiate directly from
28# the file names:
29imu2whale = imutools.IMU2Body.from_csv_nc(icsv, icdf,
30 savgol_parms=(99, 2))
31# Print summary info
32print(imu2whale)
33# Choose an index from surface details table
34idx = imu2whale.surface_details.index[33]
35idx_title = idx.strftime("%Y%m%dT%H%M%S")
IMU -- Class IMU2Body object
Source File /opt/hostedtoolcache/Python/3.12.6/x64/lib/python3.12/site-packages/skdiveMove/tests/data/gertrude/gert_imu_frame.nc
IMU: <xarray.Dataset> Size: 15MB
Dimensions: (timestamp: 171135, accelerometer: 3, magnetometer: 3,
gyroscope: 3)
Coordinates:
* timestamp (timestamp) datetime64[ns] 1MB 2017-08-10T10:07:14 ... ...
* accelerometer (accelerometer) <U1 12B 'x' 'y' 'z'
* magnetometer (magnetometer) <U1 12B 'x' 'y' 'z'
* gyroscope (gyroscope) <U1 12B 'x' 'y' 'z'
Data variables:
depth (timestamp) float64 1MB 0.0 0.0 0.0 ... 2.5 2.329 2.646
acceleration (timestamp, accelerometer) float64 4MB 0.02165 ... 0.9328
magnetic_density (timestamp, magnetometer) float64 4MB 2.259 ... -51.01
angular_velocity (timestamp, gyroscope) float64 4MB 0.7571 ... -0.3873
Attributes:
animal_id: Unknown
animal_species_common: Humpback whale
animal_species_name: Megaptera novaeangliae
deployment_id: gert01
source_files: gertrude_2017.csv,gertrude_2017_long_surfacings.csv
Surface segment duration summary:
count 68
mean 0 days 00:02:01.764705882
std 0 days 00:02:15.182918382
min 0 days 00:00:11
25% 0 days 00:00:29.750000
50% 0 days 00:00:59
75% 0 days 00:02:26.250000
max 0 days 00:08:43
dtype: object
Visualize low-pass filter (Savitzky-Golay filter;
savgol_filter()) job:
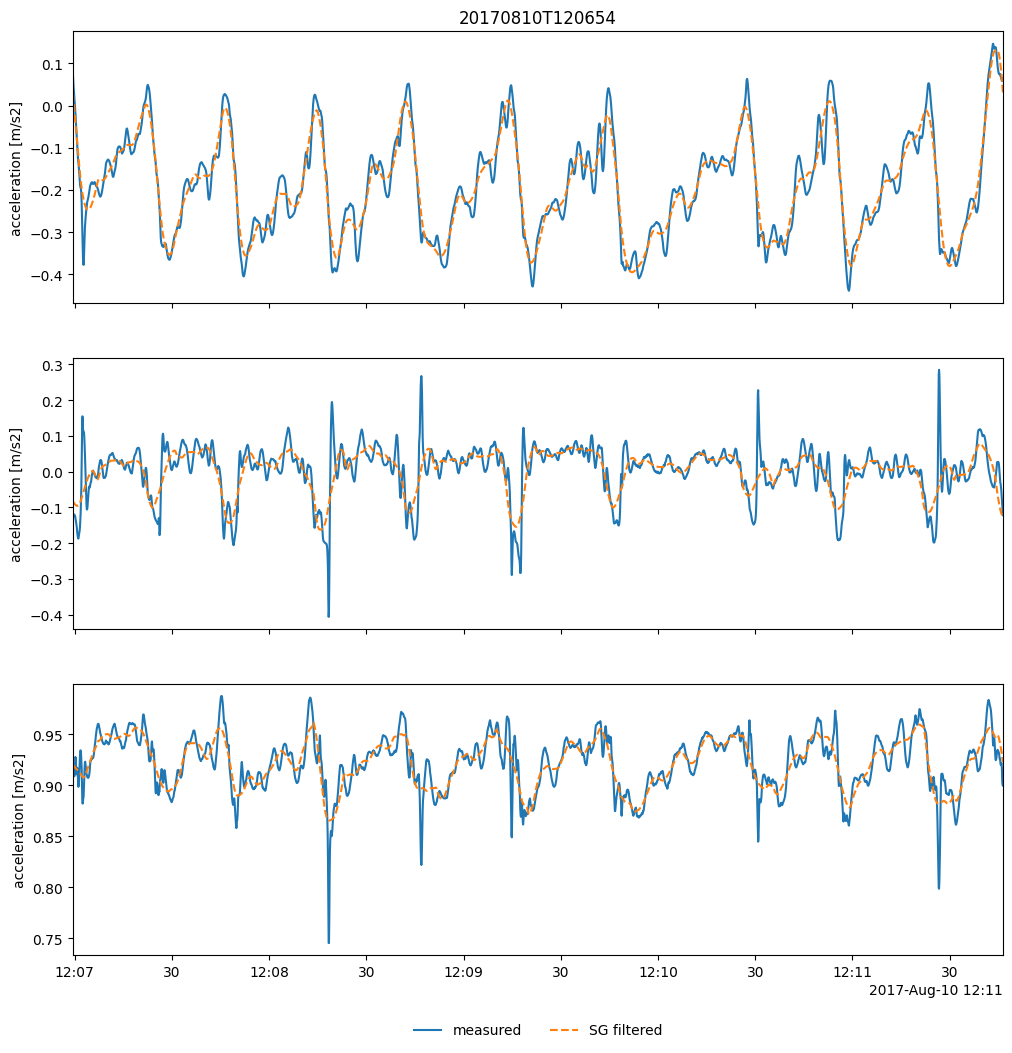
Quick plots of smoothed acceleration and magnetic density from the segment:
52acc_imu = imu2whale.get_surface_vectors(idx, _ACCEL_NAME,
53 smoothed_accel=True)
54depth = imu2whale.get_surface_vectors(idx, "depth")
55# Alternatively, use the function of the same name as method below
56ax = imu2whale.scatterIMU3D(idx, _MAGNT_NAME, normalize=True,
57 animate=False, figsize=_FIG3D1X1)
58ax.view_init(azim=-30);
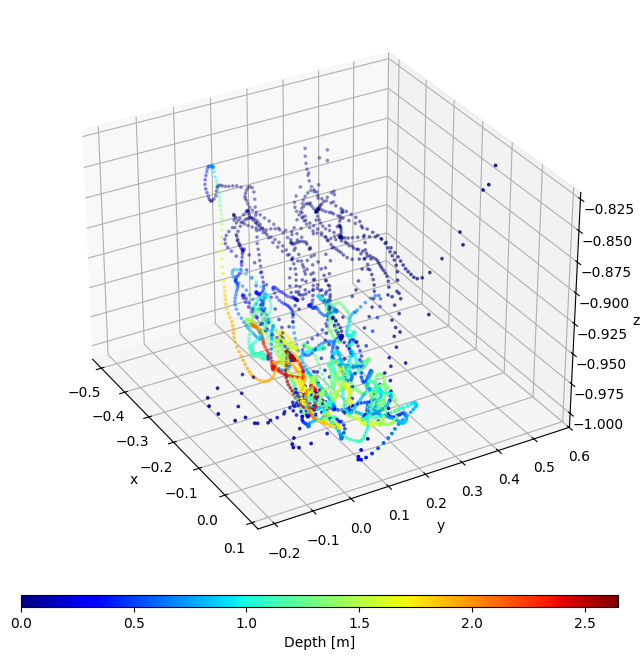
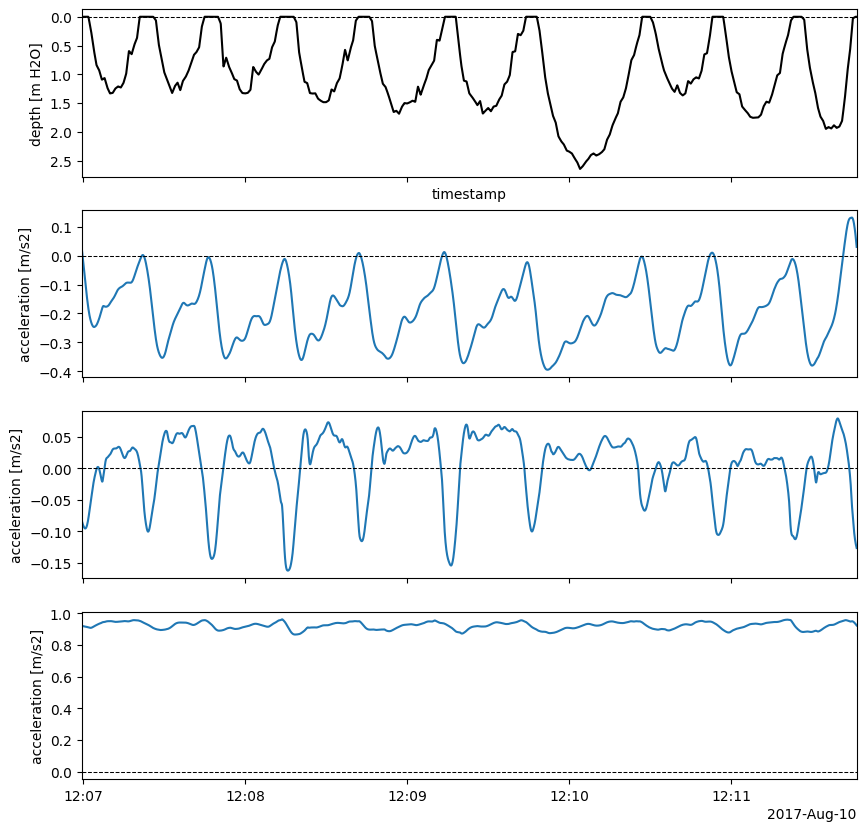
Below shows that the IMU was deployed facing forward and on the left side of the whale, so in the above plot negative x is forward and negative y is left as per our right-handed coordinate system. As above, we can use the method of the same name to produce the plot:
59imu2whale.tsplotIMU_depth(_ACCEL_NAME, idx, smoothed_accel=True,
60 figsize=_FIG4X1);
Calculate orientation for the segment above, and produce an animated plot
of the orientation. This can be done in a single step with
imutools.IMU2Body.get_orientation().
61Rctr2i, svd = imu2whale.get_orientation(idx, plot=False, animate=False)
62anim_file = "source/.static/video/gert_imu_{}.mp4".format(idx_title)
63imutools.scatterIMU_svd(acc_imu, svd, Rctr2i, normalize=True, center=True,
64 animate=True, animate_file=anim_file,
65 title="IMU-Frame Centered Acceleration [g]",
66 figsize=_FIG3D1X1)
Apply the inverse transformation to get to the animal frame:
67# Orient the surface segment using the estimated rotation
68imu_bodyi = imu2whale.orient_surfacing(idx, Rctr2i)
69# Have a look at corrected acceleration
70acci = imu_bodyi[_ACCEL_NAME]
An animation may be useful to visualize the normalized animal-frame data:
71anim_file = "source/.static/video/gert_body_{}.mp4".format(idx_title)
72imutools.scatterIMU3D(acci, imu_bodyi["depth"].to_numpy().flatten(), normalize=True,
73 animate=True, animate_file=anim_file,
74 title=r"Normalized Animal-Frame Acceleration [$g$]",
75 cbar_label="Depth [m]", figsize=_FIG3D1X1)
Obtain orientations for all surface periods, and retrieve the quality indices for each estimate:
76orientations = imu2whale.get_orientations()
77# Unpack quality of the estimates
78qual = orientations["quality"].apply(pd.Series)
79qual.columns = ["q_index", "phi_std"]
Look at the quality of the orientation estimates:
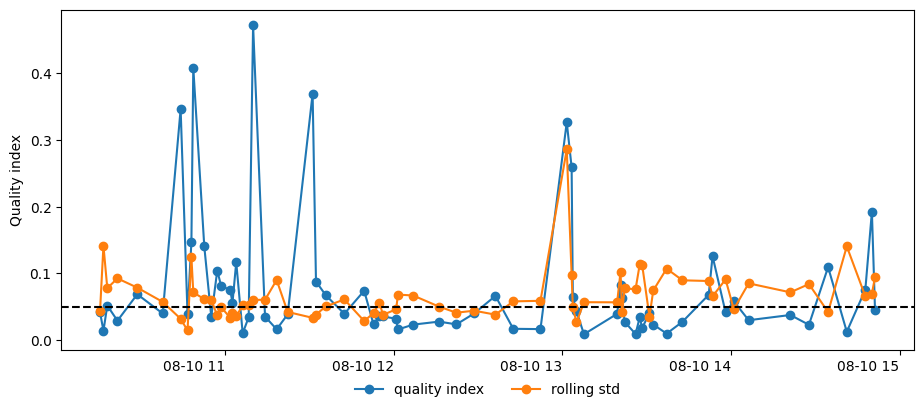
Remove bad quality estimates:
92imu2whale.filter_surfacings((0.04, 0.06))
Plot “ok” Euler angles:
| phi | theta | psi | |
|---|---|---|---|
| count | 16.000 | 16.000 | 16.000 |
| mean | -5.993 | 7.107 | -7.211 |
| std | 6.580 | 1.774 | 4.870 |
| min | -20.363 | 3.214 | -15.714 |
| 25% | -8.653 | 6.630 | -9.368 |
| 50% | -5.438 | 6.924 | -7.706 |
| 75% | -3.652 | 8.199 | -5.768 |
| max | 6.448 | 9.339 | 5.329 |
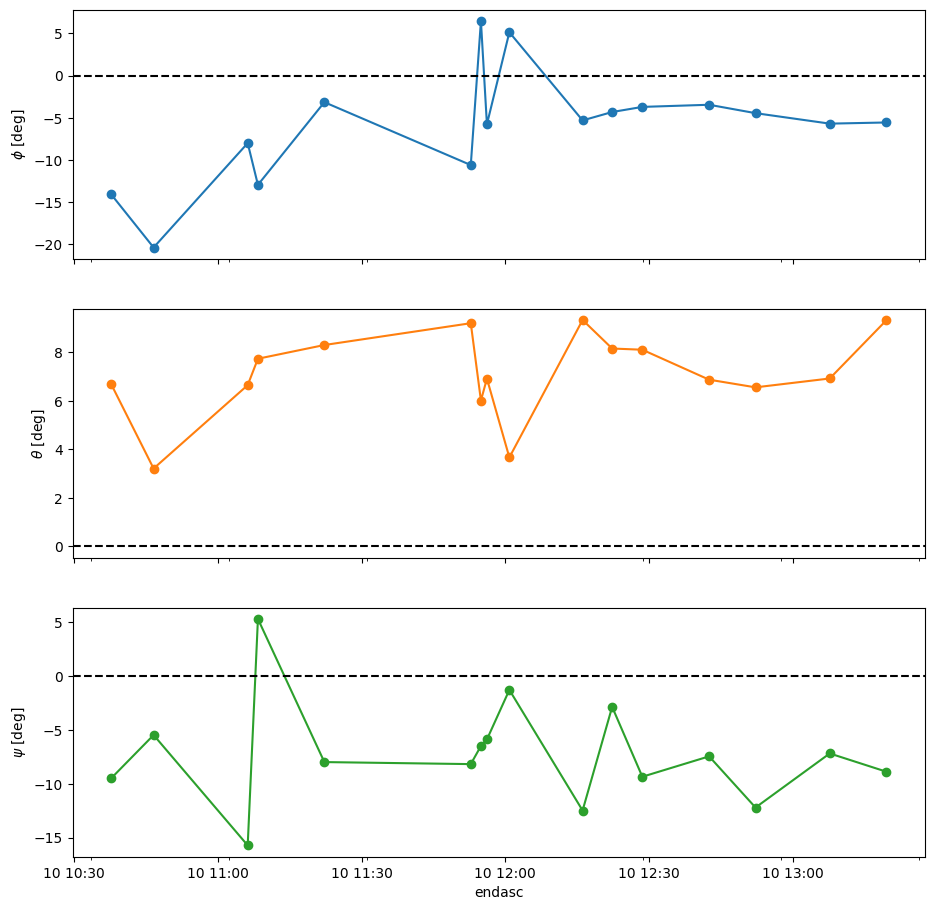
Can we use an average (median?) rotation matrix? This requires retrieving the direction cosine matrices of the centered data, which can be expressed as Euler angles with respect to the centered-data frame:
104euler_xyz = (imu2whale.orientations["R"]
105 .apply(lambda x: x.as_euler("XYZ", degrees=True))
106 .apply(pd.Series))
107euler_xyz.rename(columns={0: "phi_ctr", 1: "theta_ctr", 2: "psi_ctr"},
108 inplace=True)
109euler_avg = euler_xyz.mean()
110Rctr2i_avg = R.from_euler("XYZ", euler_avg.values, degrees=True)
111Rctr2i_avg.as_euler("XYZ", degrees=True)
112_euler_ctr2body(Rctr2i_avg)
array([-5.993, +7.107, -7.211])
Check the effect of using this common transformation with the first remaining period:
113idx0 = imu2whale.surface_details.index[0]
114imu_bodyi = imu2whale.orient_surfacing(idx0, Rctr2i_avg)
115# Plot the time series; not bad
116imutools.tsplotIMU_depth(imu_bodyi[_ACCEL_NAME], imu_bodyi["depth"],
117 figsize=_FIG4X1);
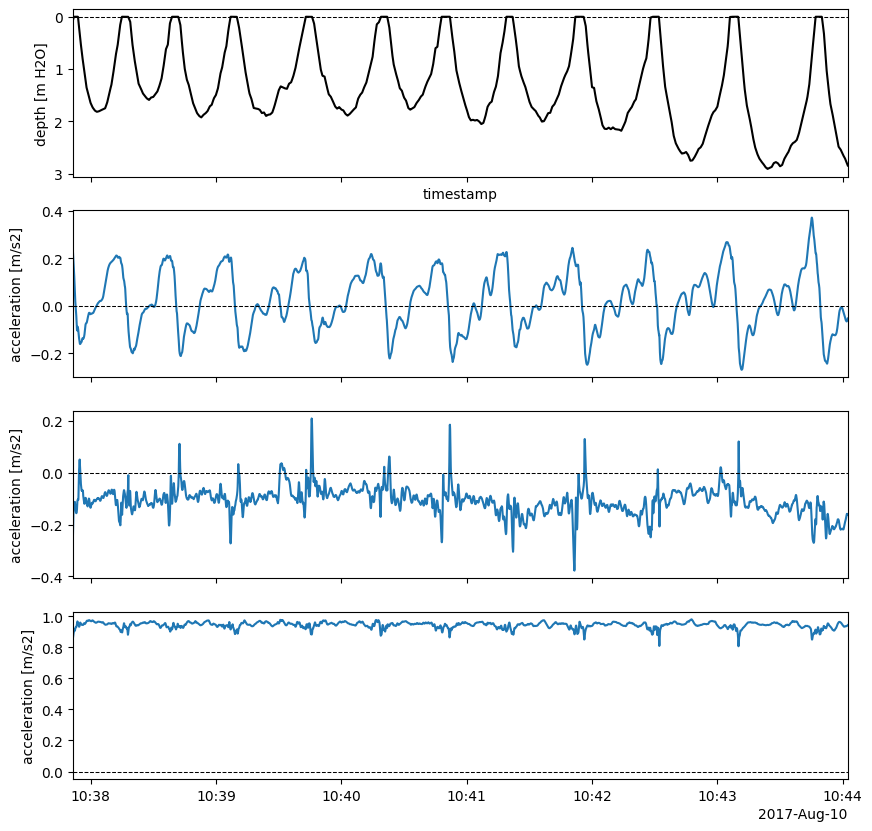
Orient all surface periods with average rotation – note we get a hierarchical dataframe output:
118imu_bodys = imu2whale.orient_surfacings(R_all=Rctr2i_avg)
119# Check out plot of a random sample
120idxs = imu_bodys.coords.get("endasc") # values in topmost level
121idx_rnd = idxs[np.random.choice(idxs.size)]
122idx_rnd_title = idx_rnd.dt.strftime("%Y%m%dT%H%M%S").item()
123# Compare with period-specific orientation
124Rctr2i = imu2whale.orientations.loc[idx_rnd.to_pandas()]["R"]
125imu_bodyi = imu2whale.orient_surfacing(idx_rnd.to_pandas(), Rctr2i)
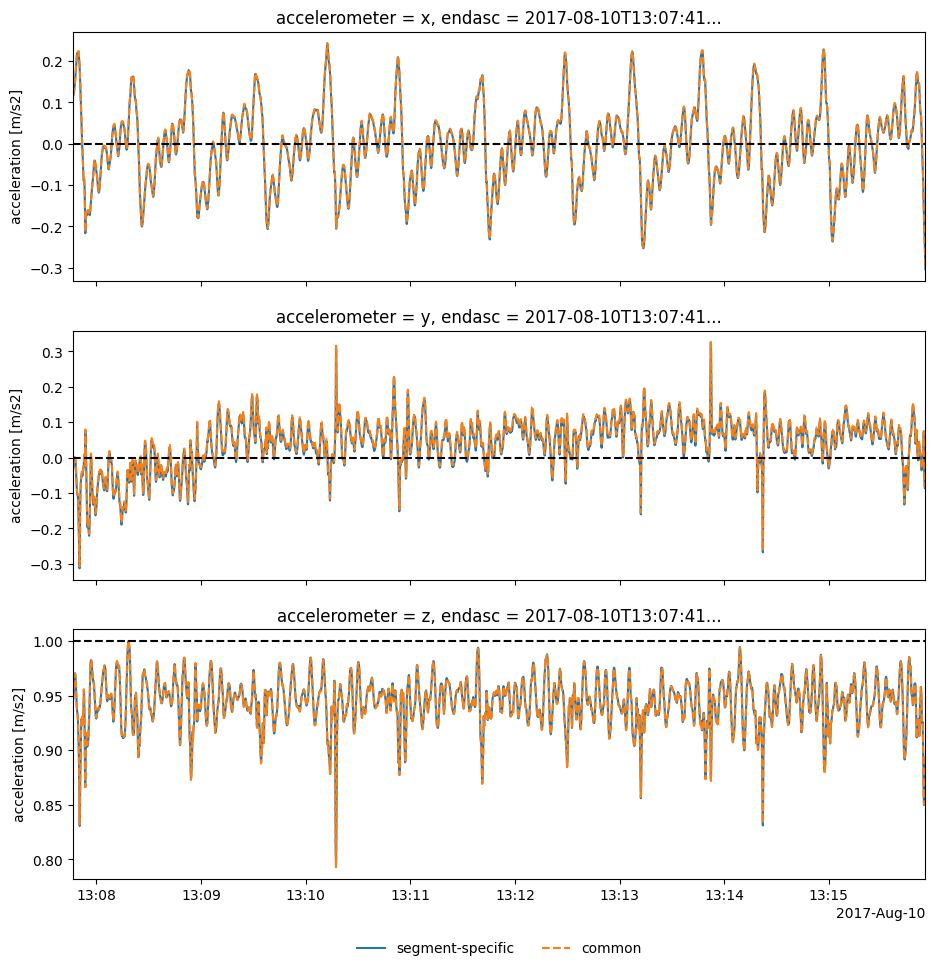
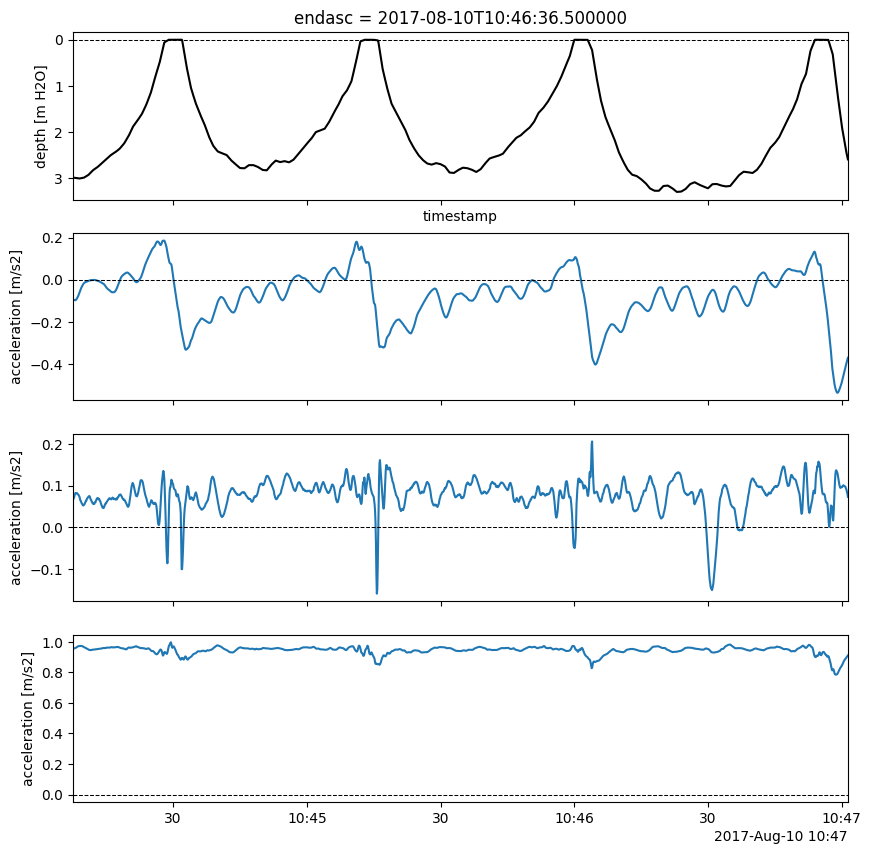
Orient the entire IMU2Body object with common rotation:
143gert_frame = imu2whale.orient_IMU(Rctr2i_avg)
Or with segment-specific rotations:
144gert_frames = imu2whale.orient_IMU()
145# Check out IMUs
146idx = imu2whale.surface_details.index[1]
147imu_i = gert_frames.sel(endasc=idx)
148imutools.tsplotIMU_depth(imu_i[_ACCEL_NAME], imu_i["depth"],
149 figsize=_FIG4X1);
Feel free to download a copy of this demo
(demo_imu2body.py).
