Simulating bouts#
This follows the simulation of mixed Poisson distributions in Luque & Guinet (2007), and the comparison of models for characterizing such distributions.
Set up the environment.
1# Set up
2import numpy as np
3import pandas as pd
4import matplotlib.pyplot as plt
5import skdiveMove.bouts as skbouts
6
7# For figure sizes
8_FIG3X1 = (9, 12)
Generate two-process mixture#
For a mixed distribution of two random Poisson processes with a mixing parameter \(p=0.7\), and density parameters \(\lambda_f=0.05\), and \(\lambda_s=0.005\), we use the random_mixexp function to generate samples.
Define the true values described above, grouping the parameters into a Series to simplify further operations.
12p_true = 0.7
13lda0_true = 0.05
14lda1_true = 0.005
15pars_true = pd.Series({"lambda0": lda0_true,
16 "lambda1": lda1_true,
17 "p": p_true})
Declare the number of simulations and the number of samples to generate:
18# Number of simulations
19nsims = 500
20# Size of each sample
21nsamp = 1000
Set up variables to accumulate simulations:
22# Set up NLS simulations
23coefs_nls = []
24# Set up MLE simulations
25coefs_mle = []
26# Fixed bounds fit 1
27p_bnd = (-2, None)
28lda0_bnd = (-5, None)
29lda1_bnd = (-10, None)
30opts1 = dict(method="L-BFGS-B",
31 bounds=(p_bnd, lda0_bnd, lda1_bnd))
32# Fixed bounds fit 2
33p_bnd = (1e-1, None)
34lda0_bnd = (1e-3, None)
35lda1_bnd = (1e-6, None)
36opts2 = dict(method="L-BFGS-B",
37 bounds=(p_bnd, lda0_bnd, lda1_bnd))
Perform the simulations in a loop, fitting the nonlinear least squares (NLS) model, and the alternative maximum likelihood (MLE) model at each iteration.
38# Set up a random number generator for efficiency
39rng = np.random.default_rng()
40# Estimate parameters `nsims` times
41for i in range(nsims):
42 x = skbouts.random_mixexp(nsamp, pars_true["p"],
43 (pars_true[["lambda0", "lambda1"]]
44 .to_numpy()), rng=rng)
45 # NLS
46 xbouts = skbouts.BoutsNLS(x, 5)
47 init_pars = xbouts.init_pars([80], plot=False)
48 coefs, _ = xbouts.fit(init_pars)
49 p_i = skbouts.bouts.calc_p(coefs)[0][0] # only one here
50 coefs_i = pd.concat([coefs.loc["lambda"], pd.Series({"p": p_i})])
51 coefs_nls.append(coefs_i.to_numpy())
52
53 # MLE
54 xbouts = skbouts.BoutsMLE(x, 5)
55 init_pars = xbouts.init_pars([80], plot=False)
56 fit1, fit2 = xbouts.fit(init_pars, fit1_opts=opts1,
57 fit2_opts=opts2)
58 coefs_mle.append(np.roll(fit2.x, -1))
Non-linear least squares (NLS)#
Collect and display NLS results from the simulations:
59nls_coefs = pd.DataFrame(np.vstack(coefs_nls),
60 columns=["lambda0", "lambda1", "p"])
61# Centrality and variance
62nls_coefs.describe()
| lambda0 | lambda1 | p | |
|---|---|---|---|
| count | 500.000 | 5.000e+02 | 500.000 |
| mean | 0.047 | 4.004e-03 | 0.729 |
| std | 0.005 | 4.165e-04 | 0.020 |
| min | 0.032 | 3.023e-03 | 0.663 |
| 25% | 0.044 | 3.715e-03 | 0.715 |
| 50% | 0.047 | 3.972e-03 | 0.729 |
| 75% | 0.050 | 4.264e-03 | 0.743 |
| max | 0.062 | 5.243e-03 | 0.785 |
Maximum likelihood estimation (MLE)#
Collect and display MLE results from the simulations:
63mle_coefs = pd.DataFrame(np.vstack(coefs_mle),
64 columns=["lambda0", "lambda1", "p"])
65# Centrality and variance
66mle_coefs.describe()
| lambda0 | lambda1 | p | |
|---|---|---|---|
| count | 500.000 | 5.000e+02 | 500.000 |
| mean | 0.050 | 5.022e-03 | 0.700 |
| std | 0.003 | 3.700e-04 | 0.022 |
| min | 0.043 | 4.047e-03 | 0.630 |
| 25% | 0.048 | 4.754e-03 | 0.684 |
| 50% | 0.050 | 5.005e-03 | 0.700 |
| 75% | 0.052 | 5.264e-03 | 0.715 |
| max | 0.059 | 6.106e-03 | 0.766 |
Comparing NLS vs MLE#
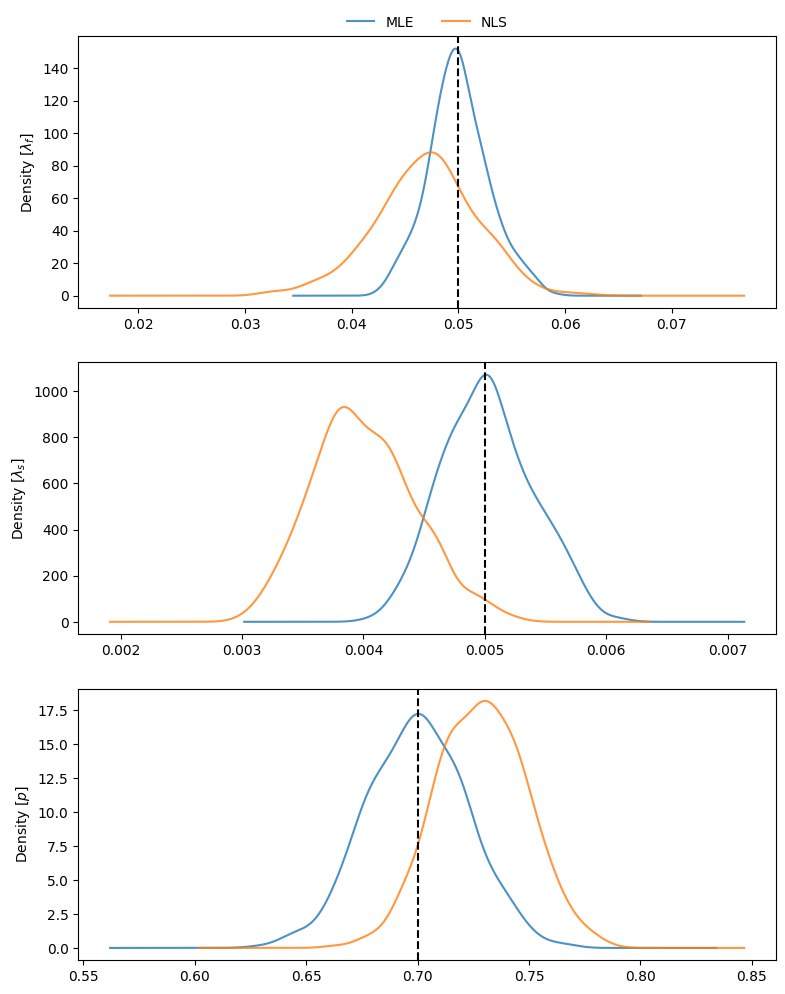
The bias relative to the true values of the mixed distribution can be readily assessed for NLS:
67nls_coefs.mean() - pars_true
lambda0 -3.029e-03
lambda1 -9.961e-04
p 2.865e-02
dtype: float64
and for MLE:
68mle_coefs.mean() - pars_true
lambda0 2.951e-05
lambda1 2.226e-05
p -1.965e-04
dtype: float64
To visualize the estimates obtained throughout the simulations, we can compare density plots, along with the true parameter values:
Three-process mixture#
We generate a mixture of “fast”, “slow”, and “very slow” processes. The probabilities considered for modeling this mixture are \(p0\) and \(p1\), representing the proportion of “fast” to “slow” events, and the proportion of “slow” to “slow” and “very slow” events, respectively.
93p_fast = 0.6
94p_svs = 0.7 # prop of slow to (slow + very slow) procs
95p_true = [p_fast, p_svs]
96lda_true = [0.05, 0.01, 8e-4]
97pars_true = pd.Series({"lambda0": lda_true[0],
98 "lambda1": lda_true[1],
99 "lambda2": lda_true[2],
100 "p0": p_true[0],
101 "p1": p_true[1]})
Mixtures with more than two processes require careful choice of constraints to avoid numerical issues to fit the models; even the NLS model may require help.
102# Bounds for NLS fit; flattened, two per process (a, lambda). Two-tuple
103# with lower and upper bounds for each parameter.
104nls_opts = dict(bounds=(
105 ([100, 1e-3, 100, 1e-3, 100, 1e-6]),
106 ([5e4, 1, 5e4, 1, 5e4, 1])))
107# Fixed bounds MLE fit 1
108p0_bnd = (-5, None)
109p1_bnd = (-5, None)
110lda0_bnd = (-6, None)
111lda1_bnd = (-8, None)
112lda2_bnd = (-12, None)
113opts1 = dict(method="L-BFGS-B",
114 bounds=(p0_bnd, p1_bnd, lda0_bnd, lda1_bnd, lda2_bnd))
115# Fixed bounds MLE fit 2
116p0_bnd = (1e-3, 9.9e-1)
117p1_bnd = (1e-3, 9.9e-1)
118lda0_bnd = (2e-2, 1e-1)
119lda1_bnd = (3e-3, 5e-2)
120lda2_bnd = (1e-5, 1e-3)
121opts2 = dict(method="L-BFGS-B",
122 bounds=(p0_bnd, p1_bnd, lda0_bnd, lda1_bnd, lda2_bnd))
123
124x = skbouts.random_mixexp(nsamp, [pars_true["p0"], pars_true["p1"]],
125 [pars_true["lambda0"], pars_true["lambda1"],
126 pars_true["lambda2"]], rng=rng)
We fit the three-process data with the two models:
127x_nls = skbouts.BoutsNLS(x, 5)
128init_pars = x_nls.init_pars([75, 220], plot=False)
129coefs, _ = x_nls.fit(init_pars, **nls_opts)
130
131x_mle = skbouts.BoutsMLE(x, 5)
132init_pars = x_mle.init_pars([75, 220], plot=False)
133fit1, fit2 = x_mle.fit(init_pars, fit1_opts=opts1,
134 fit2_opts=opts2)
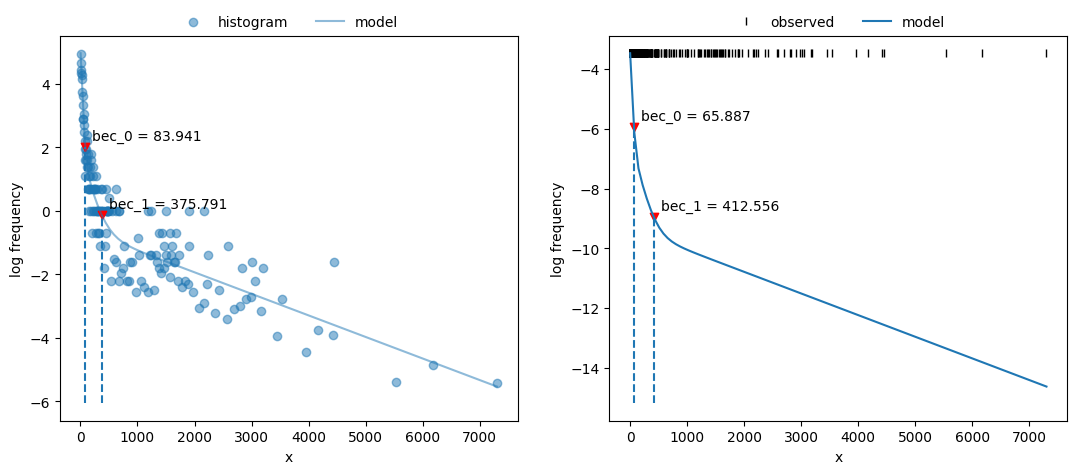
Plot both fits and BECs:
135fig, axs = plt.subplots(1, 2, figsize=(13, 5))
136x_nls.plot_fit(coefs, ax=axs[0])
137x_mle.plot_fit(fit2, ax=axs[1]);
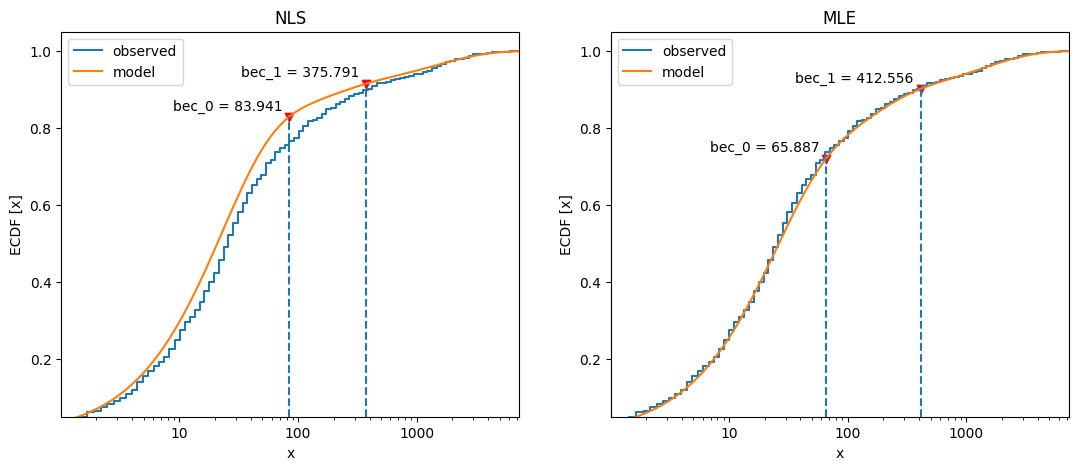
Compare cumulative frequency distributions:
138fig, axs = plt.subplots(1, 2, figsize=(13, 5))
139axs[0].set_title("NLS")
140x_nls.plot_ecdf(coefs, ax=axs[0])
141axs[1].set_title("MLE")
142x_mle.plot_ecdf(fit2, ax=axs[1]);
Feel free to download a copy of this demo
(demo_simulbouts.py).
